For 25 years, BioDiscovery, Inc. has been dedicated to developing state-of-the-art software products for life science research and clinical applications. In this interview, BioDiscovery Founder, President & CSO Soheil Shams portrays the success story of a company that is deeply entrenched in the history and rapid acceleration of genomics.
Please describe the story behind the company: What sparked the idea, and how has it evolved so far?
My background is in computer engineering and what is now called machine learning. My formal training is in AI, neural nets, and computers.
Early in my career, about 30 years ago, I got very interested in the DNA space and took a course with Mike Waterman, who is one of the earliest bioinformatics pioneers. He was teaching about DNA and RNA sequence alignment, and it made me realize the potential of merging computing with DNA and biology.
We started BioDiscovery in 1997. Trying to get into the bioinformatics field at that early stage has been a great learning experience. We’ve gone through many different phases of genomics, from initial microarrays, where people were building it themselves, to commercial arrays, to Next-generation sequencing, etc. I’m glad to say we’ve done quite well and have accumulated vast amounts of knowledge which we utilized to make our software have even greater utility in the genomics laboratory.
What can you tell us about BioDiscovery’s technology?
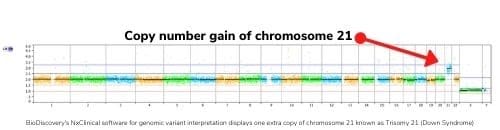
Genomics is the study of the genome, and there are many aspects to that. Our specialty so far has been on what’s called Copy Number Variation (CNVs). This is where you have an extra piece of DNA that got duplicated or deleted. A famous one is Trisomy 21 which causes Down syndrome. That’s an extra copy of chromosome 21. CNVs can be very large, like Trisomy 21, or very small. Our software also supports even smaller base level changes, known as sequence variants.
Other aspects of genomics that are very trendy these days are DNA methylation and structural variants. The latter extends CNVs to any pieces of DNA that have moved from where they were supposed to be, like translocations, insertions, deletions. I think these are fascinating areas that we’re working towards accommodating.
Our goal is to bring all of this data together: structural variants, sequence variants, CNVs, and methylation into a full picture of the patient’s genomic information.
There’s a lab in Heidelberg, Germany that’s been providing a very useful tool based on DNA methylation for classifying brain tumors, but so far, no one has effectively integrated methylation, copy number, sequence variants, and structural variance. This is where we would like to go and I would call it the leading edge of research.
What kind of clients do you typically work with?
Being in the industry for such a long time, our client mix has evolved and changed. When we first started, it was mostly research clients. We had many pharmaceutical companies, as well as research labs, especially cancer labs, such as NCI, and MD Anderson, who are now using our software, Nexus Copy Number.
Over the past decade, we have moved more towards clinical genetic testing labs and these clients are using our software, NxClinical. Right now we’re about 50-50 between research and clinical lab customers.
What kind of challenges does BioDiscovery solve for its clients?
The most common instrumentation in genomic labs reads and gives information about the genome in a digital form, regardless of whether it’s DNA microarray or next-generation sequencing. They generate massive files with large amounts of data. We try to make sense out of the data that comes off of these instruments.
For example, research clients want to understand what are common aberrations in a population with breast cancer. They want to know the difference between a grade 1 tumor and a grade 4 tumor, in terms of changes in the genome.
Our clinical application is more about the tests and their interpretation. Our software takes all the information that comes from the testing instrument and tries to make sense of that for the clinician to better understand the causes of the condition. From there, the clinician can write an informed report on the status of that patient’s disease.
For example, a young girl had developmental issues with several different phenotypes. They ran multiple DNA tests to look for what could be the cause of that, but they weren’t able to find anything until we ran it through our software and detected a small deletion of a gene that was responsible for all the phenotypes. In that case, there was no medication, but they understood what the cause was, which in itself was beneficial to the family.
If you look at cancer cases, many of those genetic aberrations have targeted therapies. For example, if someone has a KRAS mutation, they can take a particular type of medication that’s suited for that mutation. That’s another way the software helps.
While some test providers offer complex, detailed reports, others oversimplify the results. How do you balance this equation?
It’s a very good question because there is some level of possible confusion. Some companies use artificial intelligence and just generate the report with everything in it, but the field is not at the point where the machine can make those final reports.
Our tool enables the lab director, who’s a professional, to look through the data and discriminate between benign variants and things that are reportable within the context of the test. We provide all the information and the tools, but at the end of the day, the genomicist is the one who writes the report and sends it to the clinician.
What are the benefits of genetic testing for healthy consumers?
Right now, a lot of companies offer cancer predisposition tests to look at certain genes that can predispose someone, for example, to have colon cancer. Those tests are preventative in the sense that if you know you’re likely to have a particular condition, you can take early action and lower your chances of getting ill.
I did my 23andMe test very early on, about 5-6 years ago. It gave me a high-risk score for a particular disease, but the odds ratio was vague. Regulation is always an issue because you don’t want to say something if there hasn’t been enough research about it. I think they’ve gotten much better at doing that.
As far as diet and fitness recommendations go, I think up to now it has had a low impact but they are slowly becoming better and more accurate about that as well.
How do you envision the future of genomics?
I wouldn’t be in this field if I didn’t think it had a very bright future. When you look at how quickly information about the genome is coming out, you realize how revolutionary it is. I think over the next decade, a lot of medicine is going to be driven by the genomics of the patient, so I’m very excited to be part of this healthcare revolution.

